Highlights
- We compared boot covers to grab soil sampling methods in commercial fields.
- Boot covers were at least as good as soil grabs at recovering indicator microbes.
- A total of 89% of phyla and 87% of genera identified were shared by all sampling methods.
- Relative abundance of phyla and genera were highly correlated between groups.
- The phyla recovered included those containing common foodborne pathogens.
Abstract
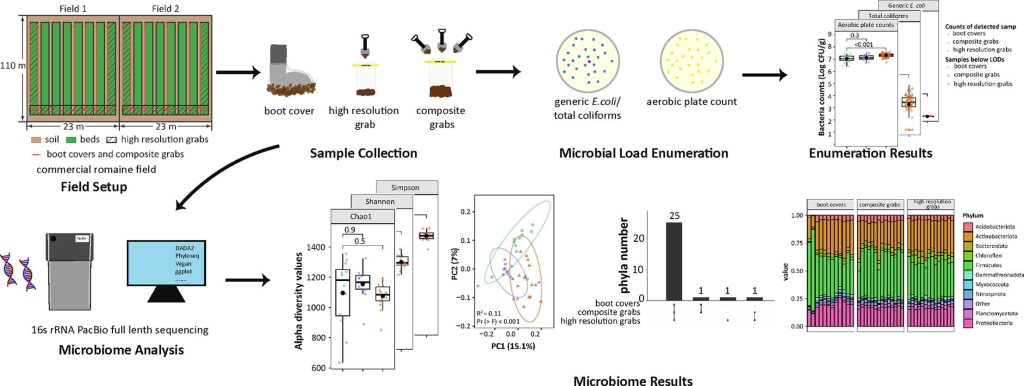
Aggregative boot cover sampling may be a more representative, practical, and powerful method for preharvest produce soil testing than grab sampling because boot covers aggregate soil from larger areas. Our study tests if boot cover sampling results reflect quality and safety indicator organisms and community diversity of grab sampling. We collected soil samples from commercial romaine lettuce fields spanning 5060 m2 using boot covers (n = 28, m = 1.1 ± 0.4 g; wearing boot covers and walking along the path), composite grabs (n = 28, m = 231 ± 24 g; consisting of 60 grabs of 3–5 g each), and high-resolution grabs (n = 72, m = 56 ± 4 g; taking one sample per stratum). Means and standard deviations of log-transformed aerobic plate counts (APCs) were 7.0 ± 0.3, 7.1 ± 0.2, and 7.3 ± 0.2 log(CFU/g) for boot covers, composite grabs, and high-resolution grabs, respectively. APCs did not show biologically meaningful differences between sample types. Boot covers recovered on average 0.6 log(CFU/g) more total coliforms than both grabs (p < 0.001) where means and standard deviations of log-transformed counts were 3.2 ± 1.0, 2.6 ± 0.6, and 2.6 ± 1.0 log(CFU/g) for boot covers, composite grabs, and high-resolution grabs, respectively. There were no generic E. coli detected in any sample by enumeration methods with LODs of 1.3–2.1 log(CFU/g) for boot covers and 0.5 log(CFU/g) for both grabs. By 16S rRNA sequencing, community species diversity (alpha diversity) was not significantly different within collection methods. While communities differed (p < 0.001) between soil sampling methods (beta diversity), variance in microbial communities was not significantly different. Of the 28 phyla and 297 genera detected, 25 phyla (89%) and 258 genera (87%) were found by all methods. Overall, aggregative boot cover sampling is similar to both grab methods for recovering quality and safety indicator organisms and representative microbiomes. This justifies future work testing aggregative soil sampling for foodborne pathogen detection.